Google’s DeepMind AI cracks 3D structure of nearly all proteins known to science in major breakthrough
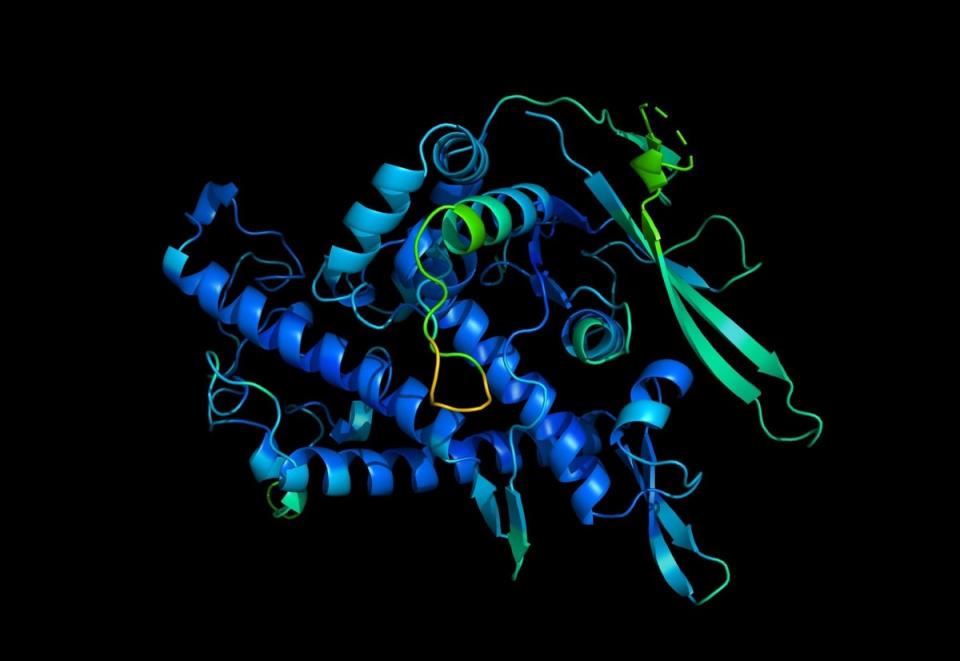
Google’s DeepMind AI has predicted the 3D structure of nearly all proteins known to science, an advance that can lead to a better understanding of rare genetic diseases, and also help develop new vaccines and drugs.
DeepMind announced on Thursday that its AlphaFold AI has cracked the structure of over 200 million proteins – the entire “universe of proteins” known to scientists.
Proteins are the building blocks of life and play myraid roles in the body as structural units, transport molecules, as well as functional catalysts of chemical reactions in the body as enzymes.
The unique 3D structure that each of these proteins takes in the body, by the folding of their constituent amino acid molecule chains, plays a major role in their function.
For decades, biologists have attempted to predict protein structures via expensive experimental means, including the use of painstaking time-consuming methods like X-ray crystallography or electron microscopy.
With the advent of computers, researchers have built virtual models of how amino acid chains making up proteins would fold under different conditions and lead to the overall 3D structure of proteins.
With the release of AlphaFold in 2020, over half a million researchers across the world have used the AI application to crack the structure of “nearly all catalogued proteins known to science.”
AlphaFold was exposed to about 100,000 known protein folding structures – already cracked by scientists – from which the AI has learned to decode the rest, the company says.
The latest advance, according to DeepMind, will expand the AlphaFold Protein Structure Database (AlphaFold DB) from nearly 1 million structures to over 200 million structures, with the potential to accelerate progress on important real-world problems “ranging from plastic pollution to antibiotic resistance.”
In the new update, DeepMind has added the predicted structures for proteins found in plants, bacteria, animals, and other organisms, which may help solve important global issues, “including sustainability, food insecurity, and neglected diseases,” the company noted in a statement.
“You can think of it as covering the entire protein universe. We’re at the beginning of a new era now in digital biology,” DeepMind chief Demis Hassabis said at a press briefing.
A big day for #AI in life science. Release of >200 million predicted 3D protein structures from open-source #AlphaFold, nearly the entire protein universe
See: https://t.co/gjASHqACqa @DeepMind
my comment below pic.twitter.com/yPgtPHMZac— Eric Topol (@EricTopol) July 28, 2022
With the new structure predictions, scientists can better understand if variant forms of the proteins that differ between individuals are linked to diseases.
For example, protein structures predicted by AlphaFold are helping in the development of drugs for neglected tropical diseases like leishmaniasis and Chagas disease – illnesses that disproportionately affect people in poorer parts of the world.
And in April, scientists at the Yale University used AlphaFold’s database to develop a new Malaria vaccine.
By cracking the structure of key proteins in the body, linked to diseases, scientists can model drugs that can effectively activate or take the role of malfunctioning proteins, or suppress those causing problems.
Decoding protein structures do not just aid in curing diseases but can also help engineer solutions for global environmental issues.
For instance, researchers have joined hands with DeepMind’s AI to develop faster-acting enzymes to break down and recycle some of the world’s most polluting single-use plastics.
“AlphaFold is the singular and momentous advance in life science that demonstrates the power of AI. Determining the 3D structure of a protein used to take many months or years, it now takes seconds,” Eric Topol, Founder and Director of the Scripps Research Translational Institute, said.
“AlphaFold has already accelerated and enabled massive discoveries, including cracking the structure of the nuclear pore complex. And with this new addition of structures illuminating nearly the entire protein universe, we can expect more biological mysteries to be solved each day,” Dr Topol added.
 Yahoo Finance
Yahoo Finance 